Group of Integrated Genomic Breeding
Faculty staff
Prof. Dr. Toshio Yamamoto
E-mail: yamamo101040ATokayama-u.ac.jp
(please change AT to @ before sending)

Assoc. Prof. Dr. Kiyotaka Nagaki
E-mail: nagakiATokayama-u.ac.jp
(please change AT to @ before sending)

Assist. Prof. Dr. Tomoyuki Furuta
E-mail: f.tomoyukiATokayama-u.ac.jp
(please change AT to @ before sending)

Summary of main research topics
For other topics, please see the lab web page.
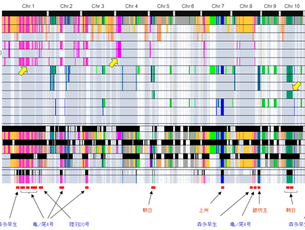
1. Exploring and utilization of useful traits from diverse rice genetic resources
Rice is cultivated all around the world and shows numerous morphological and physiological differences in the form of phenotypic variations. Some of these variations have been used as genetic resources to improve rice plants so that they better satisfy human needs. Phenotypic variations are considered to be genetically controlled by the collective function of a large number of genes on rice genomes However, the genetic bases and biological functions of most of them are still unknown, which has prevented us from wider practical application of rice germplasms. We exploit useful phenotypic variations from a wide range of rice germplasms and clarify the genes involved with biological functions by aid of recent advances in genome information and technology. Also, we try to develop new breeding materials and propose more effective breeding methodologies.
2. Development of remote cross breeding in rice via polyploidization
For breeding of super rice varieties with useful new genes, it is important to promote allelic exchange in existing cultivars by crossing them with wild or genetically remote cultivars. However, hybrids are rarely obtained from such distant crosses due to multitude of reproductive barriers. Interestingly, we found a fertile tetraploid progeny derived from anther culture of Asian rice cultivar, O. sativa, and African rice cultivar, O. glaberrima. Using genomic and phenotypic analyses, we now aim to clarify mechanisms involved in recovery of seed set in these plants. Furthermore, we hope to establish a novel remote cross breeding strategy in rice that overcomes reproductive barriers by introduction of polyploidization and haploidization.

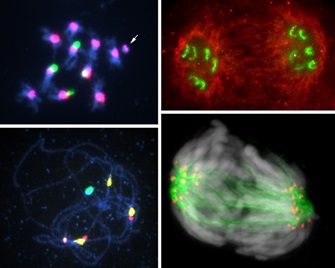
3. Design and synthesis of ‘chromosomes’ carrying new genome information
Nuclei that have very complex structures and various functions are the most important organelles in eukaryotic cells. Nuclear DNA are divided and packed into chromosomes, enabling the accurate transmission of genetic information to daughter cells. Our research group is studying the molecular structures and functions of nuclei and chromosomes, mainly in plants. Our most recent goal is the development of plant artificial chromosomes to elucidate chromosome functional elements: centromeres, telomeres, and replication origins. We are also interested in the relation between chromatin modifications and gene expression.
4. Boosting up crop breeding by integrated bioinformatics and statistical genetics
Along with drastically changing our lives, computers have also brought a paradigm shift in plant breeding. “Genomic breeding” is a state-of-the-art method which integrates genome-wide genotype information to model and predict phenotypic variation in populations. Although modern computer-based methods have strong potential for accelerating the development of new varieties, statistical models used in the method still need improvements to capture in detail events happening in real nature. At present, we try to integrate all useful environmental and biological data into new statistical models for genomic breeding that will allow accurate phenotype prediction within relatively small datasets. We apply a large variety of modeling methods that also include machine learning strategies. We hope that our work will boost up breeding in the future, helping to overcome the global shortage in food production experienced worldwide.
Publication List
(1) Samadi, A. F., Suzuki, H., Ueda, T., Yamamoto, T., Adachi, S. and Ookawa, T. Identification of quantitative trait loci for breaking and bending types lodging resistance in rice, using recombinant inbred lines derived from Koshihikari and a strong culm variety, Leaf Star. Plant Growth Regulation 89(1): 83-98. (2019. 1.)
(2) Sun, J., Ma, D., Tang, L., Zhao, M., Zhang, G., Wang, W., Song, J., Li, X., Liu, Z., Zhang, W., Xu, Q., Zhou, Y., Wu, J., Yamamoto, T., Dai, F., Li, S., Zhou, G., Zheng, H., Xu, Z. and Chen, W. Population Genomic Analysis and De novo Assembly Reveal the Origin of Weedy Rice as an Evolutionary Game. Mol. Plant 12(5): 632-647. (2019. 5.)
(3) Adachi, S., Yamamoto, T., Nakae, T., Yamashita, M., Uchida, M., Karimata, R., Ichihara, N., Soda, K., Ochiai, T., Ao, R., Otsuka, C., Nakano, R., Takai, T., Ikka, T., Kondo, K., Ueda, T., Ookawa, T. and Hirasawa, T. Genetic architecture of leaf photosynthesis in rice revealed by different types of reciprocal mapping populations. Journal of Experimental Botany 70(19): 5131-5144. (2019. 6.)
(4) Nomura, T., Arakawa, N., Yamamoto, T., Ueda, T., Adachi, S., Yonemaru, J., Abe, A., Takagi, H. and Ookawa, T. Next generation long-culm rice with superior lodging resistance and high grain yield, Monster Rice 1. PLoS ONE 14(8): e0221424. (2019. 8.)
(5) Ogawa, D., Sakamoto, T., Tsunematsu, H., Yamamoto, T., Kanno, N., Nonoue, Y. and Yonemaru, J. Surveillance of panicle positions by unmanned aerial vehicle to reveal morphological features of rice. PLoS ONE 14(10): e0224386. (2019. 10.)
(6) Kuya, N., Sun, J., Iijima, K., Venuprasad, R. and Yamamoto, T. Novel method for evaluation of anaerobic germination in rice and its application to diverse genetic collections. Breeding Science https://doi.org/10.1270/jsbbs.19003 (2019. 10. Online preview)
(7) San, N. S., Suzuki, K., Soda, K., Adachi, S., Kasahara, H., Yamamoto, T., Ikka, T., Kondo, K., Yamanouchi, U., Sugimoto, K., Nagamura, Y., Hirasawa, T. and Ookawa, T. Semi-dwarf 1 (sd1) gene enhances light penetration into the canopy through regulating leaf inclination angle in rice. Field Crops Research https://doi.org/10.1016/j.fcr.2019.107694 (2019. 12. Online preview)