Germplasm Center
BARLEY
Barley
Barley is the fourth major cereal crop in the world. About 10,000 years ago it was domesticated from a wild ancestor in the Fertile Crescent and now it is grown throughout the world excepting the tropics because of its wide adaptability to the salt, drought, low temperature etc. The barley varieties show a large variation in morphological, ecological and agricultural traits such as plant type, ear type, growth habit, maturity, plant height, grain size, disease resistance etc. About 3,000 years ago barley was introduced to Japan from the continent, and it used to be an important food crop. Barley was cooked with rice when the rice production was limited, and sometimes we eat barley in this way still now. At present, barley is used mainly as feed of livestocks, malts for brewing beer and whisky, miso and Mugicha-tea.
History of Barley Germplasm Center
Barley collection started in early 1940’s by Dr. Ryuhei Takahashi for the crop evolution and domestication study. The collection expanded according to the detailed study of traits related to evolution and breeding. In 1979 the Ministry of Education, Science, Sports and Culture established the Barley Germplasm Center at the Institute for Agricultural and Biological Sciences (renamed as Research Institute for Bioresources in 1989, Institute of Plant Science and Resources in 2010), Okayama University located at Kurashiki. Currently, Barley Germplasm Center is one of the main international barley genebanks and supported by National Bioresource Project (http://www.nbrp.jp/) for the preservation and distribution
Functions of Barley Germplasm Center
The functions of the Center are to (1) collect or develop materials (2) preserve materials (3) evaluate and genetically analyze the materials, (4) construct databases and (5) distribute seed samples upon request.
Collection
We have preserved ca. 14,000 accessions of cultivated barley including experimental lines and ca. 600 accessions of wild relatives.
Preservation
Barley is usually sown in November and harvested until early June in Kurashiki where the climate is mild in winter but has rainy season from June. Each accession is checked the recorded marker traits i.e. plant type, hairiness of leaf sheath, heading time, ear type, awn length, hull type. Then barley spikes are harvested, dried and threshed very carefully to prevent the contamination. After fumigated with chemical insecticide the seeds are stored in storage room at 15℃ (short and middle term) or -30℃ (long term) under dry condition.
Evaluation
Resources
Requesting resources
Resources on the databases http://www.shigen.nig.ac.jp/barley/ can be distributed. Please check databases and contact us by email on the web page. MTA must be signed before shipment. Cost for handling and shipment is charged.
Collection
Barley seed samples
Ca. 6000 accessions on the database are distributed.
ESTs
134,928 entries are published and their clones are available
FLcDNAs
5,006 entries are published and their clones are available
BACs
Harina Nijo BAC library: A set of clones (756 x 384 MTPs), 16 large filters, Plate pooled DNAs, Super-pooled DNAs are available
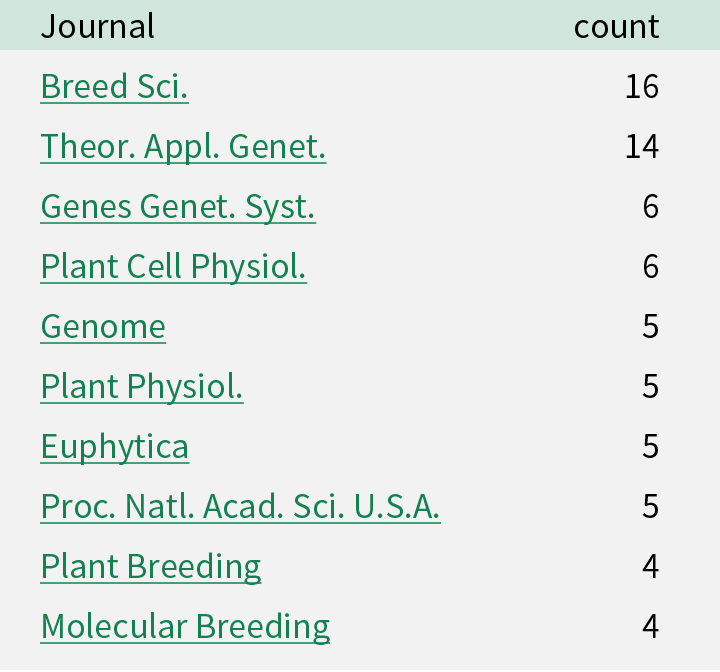
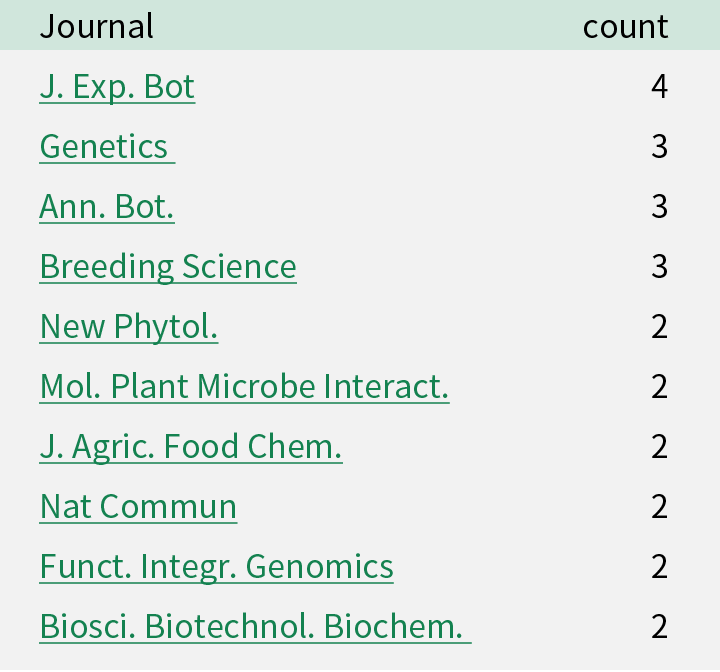
Publications using barley resources
Following papers have been published during the project. Please visit the list at http://www.shigen.nig.ac.jp/rrc/index.jsp
Genome Analysis
The project ‘Identification of genes of important traits and their application in barley breeding’ has just finished with support of Bio-oriented Technology Research Advancement Institution (BRAIN). The project aimed to sequence genes on chromosome 3H and isolate genes responsible for brewing traits and stress tolerances. The full length cDNA projects on barley are also conducted by National Bioresource Project and Genome diversity analysis project by MAFF. Please visit Databases page for sequence data.
Items
Sequencing
Currently, barley genome is sequenced by the international barley sequencing consortium using next generation sequencers.
cDNA collections and applications
135,000ESTs and 5,000 FLcDNAs have been sequenced and published at databases. Of these, 80,000 clones were distributed in NBRP project. GeneChip Barley 1 Array was developed from EST sequences.
BAC clones
A BAC library by Japanese cultivar was developed and distributed. Membrane screening system (16 large filters) and DNA plate pools for PCR screening are also distributed.
EST maps
A high density transcript map was developed and published as an electornic map (cMAP).
Comparative analysis
A diploid wheat genetic map have been published in paper and database.
Databases
Databases and Genome tools
Barley germplasm
Barley EST/FLcDNA
Mapping information of barley FLcDNA on rice genome
Electronic map of 3K barley 3′ ESTs
Barley genome sequences


