
Research
Group of Integrated Genomic Breeding
| Link to group homepage |
Faculty staff
 |
Prof. Dr. Toshio Yamamoto E-mail: yamamo101040ATokayama-u.ac.jp |
(please change AT to @ before sending) | ||
 |
Assoc. Prof. Dr. Kiyotaka Nagaki E-mail: nagakiATokayama-u.ac.jp |
(please change AT to @ before sending) | ||
 |
Assoc. Prof. Dr. Tomoyuki Furuta E-mail: f.tomoyukiATokayama-u.ac.jp |
(please change AT to @ before sending) |
Lectures: Analytical Molecular Cytogenetics
Keywords: Centromere; Artificial chromosome; Nucleus; Chromatin
Summary of main research topics
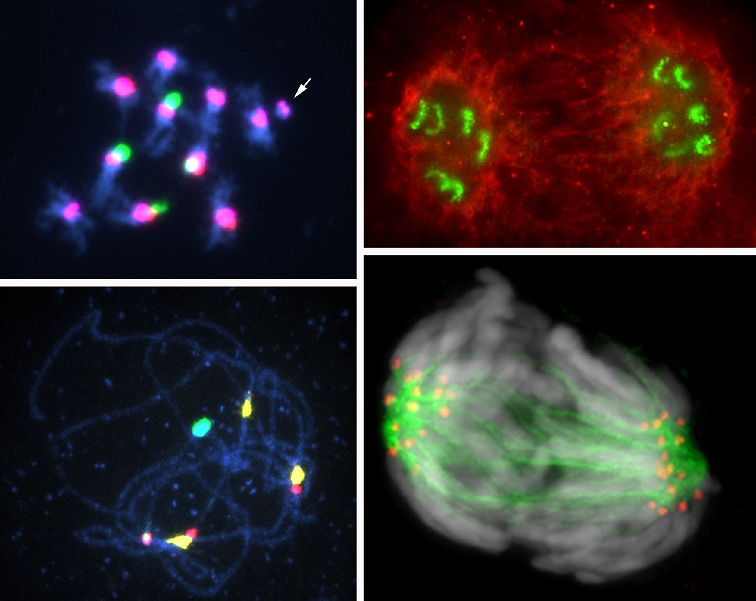
| Exploring and utilization of useful traits from diverse rice genetic resources Rice is cultivated all around the world and shows numerous morphological and physiological differences in the form of phenotypic variations. Some of these variations have been used as genetic resources to improve rice plants so that they better satisfy human needs. Phenotypic variations are considered to be genetically controlled by the collective function of a large number of genes on rice genomes However, the genetic bases and biological functions of most of them are still unknown, which has prevented us from wider practical application of rice germplasms. We exploit useful phenotypic variations from a wide range of rice germplasms and clarify the genes involved with biological functions by aid of recent advances in genome information and technology. Also, we try to develop new breeding materials and propose more effective breeding methodologies. Design and synthesis of ‘chromosomes’ carrying new genome information Nuclei that have very complex structures and various functions are the most important organelles in eukaryotic cells. Nuclear DNA are divided and packed into chromosomes, enabling the accurate transmission of genetic information to daughter cells. Our research group is studying the molecular structures and functions of nuclei and chromosomes, mainly in plants. Our most recent goal is the development of plant artificial chromosomes to elucidate chromosome functional elements: centromeres, telomeres, and replication origins. We are also interested in the relation between chromatin modifications and gene expression. |
 |
Latest publications (for complete and most current publications visit group pages)
(1) Furuta T, Saw O.M, Moe S, Win K.T, Hlaing M.M, Hlaing A.L.L, Thein M.S, Yasui H, Ashikari M, Yoshimura A, Yamagata Y. Development of genomic and genetic resources facilitating molecular genetic studies on untapped Myanmar rice germplasms. Breed. Sci. 74: 124-137. doi.org/10.1270/jsbbs.23077 (2024. 4.)
(2) Hlaing M.M, Yamagata Y, Furuta T, Win K.T, Saw O.M, Ozaki A, Yasui H, Yoshimura A. Genetic variation in heading dates and phenological parameters of Myanmar rice. Plant Prod. Sci. 27: 125-136. doi.org/10.1080/1343943X.2024.2308336 (2024. 4.)
(3) Yamamoto T, Kashihara K, Furuta T, Zhang Q, Yu E, Ma J.F. Genetic background influences mineral accumulation in rice straw and grains under different soil pH conditions. Sci. Rep. 14: 15139. doi.org/10.1038/s41598-024-66036-7 (2024.7.)
(4) Toriyama K, Iwai Y, Takeda S, Takatsuka A, Igarashi K, Furuta T, Chen S, Kanaoka Y, Kishima Y, Arimura Si, Kazama T. Cryptic cytoplasmic male sterility-causing gene in the mitochondrial genome of common japonica rice. Plant J. 120: 941-949. doi.org/10.1111/tpj.17028 (2024. 9.)
(5) Tek A.L, Nagaki K, Akkamis Y.H, Tanaka K, Kobayashi H. Chromosome-specific barcode system with centromeric repeat in cultivated soybean and wild progenitor. Life Sci. Alliance. 7: 10. doi.org/10.26508/lsa.202402802 (2024.10.)





